Baseline-ZERO DNase
Key features
Show- Effectively digest large or small dsDNA and ssDNA into mononucleotides
- Digest unwanted dsDNA and ssDNA molecules including small ssDNA such as random hexamers
- Use in sensitive applications requiring reverse transcription where any contaminating DNA is unwanted
Size: 5000 units
Product information
Baseline-ZERO™ DNase* digests dsDNA and ssDNA into mononucleotides more effectively than the commonly used bovine pancreatic DNase I. Even the small DNA oligonucleotides that remain after treatment with bovine pancreatic DNase I are undetectable by gel electrophoresis following treatment with Baseline-ZERO DNase (Figure. 2). Removal of DNA from RNA preparations is particularly beneficial when RNA in a sample is amplified using a method that involves reverse transcription using random primers, since any contaminating DNA would also be a template for random-primed cDNA synthesis.
Applications
- Removal of genomic DNA from small-sample total RNA preparations for expression analysis (Figure. 1)
- Removal of small DNA oligonucleotides (e.g. random primers)
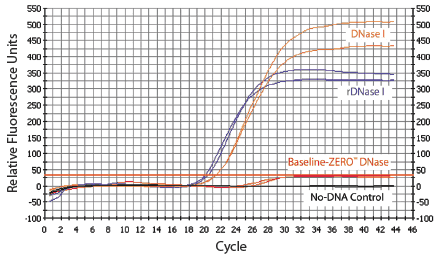
Figure 1. Real-time PCR of HeLa RNA preparations treated with various DNases. The lower the C T value (intersection of curves with the red line), the greater the amount of residual DNA not digested by the indicated DNase. Thus, Baseline-ZERO™ DNase removed all detectable DNA from the RNA sample. The TaqMan® probe assay amplified a 268-bp fragment of β-actin. Samples were run in duplicate.

References
- Kienzle, N. et al. (1996) BioTechniques 20, 612.
*Patent pending.
Unit Definition: One Molecular Biology Unit (MBU) of Baseline-ZERO™ DNase produces an increase in the A260 of a solution of dsDNA, of 0.001 per minute at 25 °C. Functionally, 1 MBU completely digests 1 µg of linear pUC19 DNA to mononucleotides in 10 minutes at 37 °C.
Storage Buffer: Baseline-ZERO DNase is supplied in a 50% glycerol solution containing 50 mM Tris-HCl (pH 7.5), 10 mM CaCl2, 10 mM MgCl2 and 0.1% Triton® X-100.
10X Baseline-ZERO™ DNase Reaction Buffer: 100 mM Tris HCl (pH 7.5), 25 mM MgCl2 and 5 mM CaCl2.
10X Baseline-ZERO™ DNase Stop Solution: 30 mM EDTA.
Quality Control: Baseline-ZERO DNase is assayed for its ability to completely digest linear dsDNA to mononucleotides under standard assay conditions. Baseline-ZERO DNase is free of detectable RNase activities as assayed by PAGE analysis of 1 µg of a synthetic RNA transcript following an overnight incubation with sufficient DNase I to completely digest 1,000 µg of DNA.
SDS
Manuals and user guides
Product information sheets
Access support
Need some support with placing an order, setting up an account, or finding the right protocol?
Contact us