DNA and RNA Purification Kits and Reagents
MasterPure Yeast DNA Purification Kit
Product Details
ShowContents: Yeast Cell Lysis Solution, MPC Protein Precipitation Reagent, TE Buffer, RNase A.
MasterPure Yeast DNA Purification Kit
Key features
Show- Obtain high yields of quality DNA from yeast with this easy to use kit
- Fast: Obtain purified DNA in less than 40 minutes
- Flexible: Purify DNA from a wide variety of yeast species, including Candida, Saccharomyces, Pichia, and Schizosaccharomyces, and filamentous fungi such as Aspergillus2 and Penicillium
- High Quality DNA: Purify high molecular weight genomic DNA that is ready to use in many molecular biology applications, including PCR amplification, restriction endonuclease digestion, Southern blotting, genomic library preparation and fungal identification and typing
- High Yields: Purify more yeast genomic DNA than other commercially available kits
- Simple: No enzymatic lysis, bead beating, columns, phenol or other organic solvents are used
Number of purifications: 200
Similar Products
DNA and RNA Purification Kits and Reagents
Tissue & Cell Lysis Solution
Product information
The MasterPure™ Yeast DNA Purification Kit enables efficient, high-yield purification of high-molecular-weight DNA from yeast and other fungi. The protocol involves non-enzymatic cell lysis at 65 °C, followed by removal of protein by precipitation, and nucleic acid precipitation and re-suspension. No lyticase, proteolytic enzymes, or bead-beating are used in the procedure. Yeast genomic DNA yields using the MasterPure Kit are much higher than yields obtained with other commercially available kits (Figure. 1). 1 The protocol can be easily adjusted for larger or smaller samples, including single yeast colonies. 1 The recovered nucleic acid can be used directly in most applications, including PCR amplification.
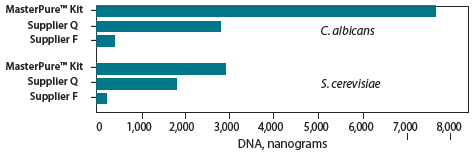
Figure 1. Comparative yields of DNA from the MasterPure Yeast DNA Purification Kit and competitor kits. DNA was quantitated specifically with Hoechst fluorescent dye 33258, which gives minimal fluorescence with RNA. The data represent the average DNA yields determined by fluorometry from two experiments with S. cerevisiae and C. albicans. The MasterPure Kit produced up to 17 times more DNA from C. albicans and 12 times more DNA from S. cerevisiae than other kits.

Figure 2. Size distribution of DNA produced from the MasterPure Kit. For each kit indicated, a 500-ng aliquot of purified yeast DNA was analysed by pulse-field gel electrophoresis on a 1% agarose gel. The gel was stained with ethidium bromide. The majority of DNA isolated with the MasterPure Kit was estimated to be in the 40- to 50-kb size range, while degradation to smaller fragments was observed with other kits. Lane M, lambda DNA ladder; Lane 4, T7 DNA (40 kb).
References
- Hoffman, L. and Moan, E. (1998) Epicentre Forum 5(4), 1.
- Jin, J., Lee, Y.-K. and Wickes, B.L. (2004) J. Clin Microbiol. 42, 4293.
SDS
Manuals and user guides
Access support
Need some support with placing an order, setting up an account, or finding the right protocol?
Contact us